基于分子形状的比对对于虚拟筛选和骨架跃迁都是非常有用的。
导入库
import os
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit import RDConfig
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem import Draw
from rdkit.Chem import PyMol
from rdkit.Chem.Subshape import SubshapeBuilder, SubshapeAligner, SubshapeObjects载入数据
mols = [ Chem.AddHs(m) for m in Chem.SDMolSupplier("cdk2.sdf")]可视化(调用Pymol)
v = PyMol.MolViewer()
cidslist=[]
for mol in mols:
mol.RemoveAllConformers()
cids = AllChem.EmbedMultipleConfs(mol, pruneRmsThresh=1.0)
cidslist.append(cids)
for idx in range(len(mols)):
mol = mols[idx]
for cid in cidslist[idx]:
AllChem.UFFOptimizeMolecule(mol, confId=cid)绘制分子
Draw.MolsToGridImage(mols[:5], molsPerRow=5)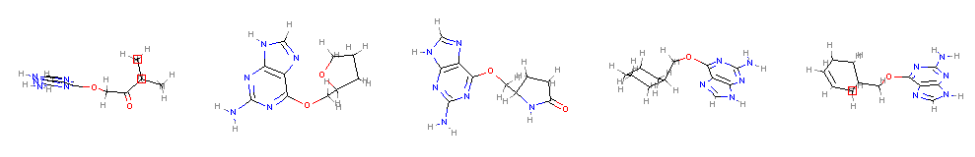
设置可视化参数
ref = Chem.Mol(mols[0].ToBinary())
probe = Chem.Mol(mols[1].ToBinary())
AllChem.CanonicalizeConformer(ref.GetConformer())
builder = SubshapeBuilder.SubshapeBuilder()
builder.gridDims = (20.,20.,10.)
builder.gridSpacing=0.5
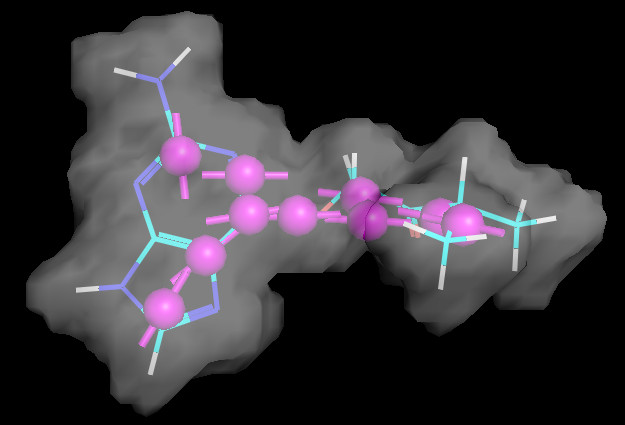
builder.winRad = 4.分子形状比对
refShape = builder.GenerateSubshapeShape(ref)
probeShape = builder.GenerateSubshapeShape(probe)可视化比对结果
v.ShowMol(ref,name="ref")
SubshapeObjects.DisplaySubshape(v,refShape,'ref_Shape')
v.server.do('set transparency=0.5')
v.GetPNG()
